[edit]
1 мҷңлҸ„(skewness) #
мҷңлҸ„лҠ” 분нҸ¬мқҳ 비лҢҖм№ӯ м •лҸ„лҘј мёЎм •н•ҳлҠ” мІҷлҸ„
skew.f<-function(x)
{
x<-as.matrix(x)
n<-nrow(x)
m<-cov.wt(x)$center
S<-cov.wt(x,method="ML")$cov
SI<-solve(S)
skew<-0
for(r in 1:n) {
for(s in 1:n) {
skew<-skew+(t(x[r,]-m)%*%SI%*%(x[s,]-m))^3
}
}
skew/(n^2)
}
[edit]
2 мІЁлҸ„(kurtosis) #
мІЁлҸ„лҠ” 분нҸ¬мқҳ мң— л¶Җ분мқҳ нҸүнҸүн•ң м •лҸ„лҘј мёЎм •н•ҳлҠ” мІҷлҸ„
kurtosis.f<-function(x)
{
x<-as.matrix(x)
n<-nrow(x)
m<-cov.wt(x)$center
S<-cov.wt(x,method="ML")$cov
SI<-solve(S)
kurtosis<-0
for(r in 1:n) {
kurtosis<-kurtosis+( t(x[r,]-m) %*% SI %*% (x[r,]-m) )^2
}
kurtosis/n
}
[edit]
3 м •к·ңм„ұ кІҖм • #
normality.f<-function(x,alpha=0.05)
{
n<-nrow(x)
p<-ncol(x)
m<-(p*(p+1)*(p+2))/6
b1<-skew.f(x)
p.val<-1-pchisq((n*b1)/6,m)
cat("Skewness is ",b1,"and P-value is ",p.val,".\n")
if(p.val < alpha)
cat("Reject the normality.\n")
else
cat("Don't reject the normality.\n")
}
- alpah : мң мқҳмҲҳмӨҖ
- m: мһҗмң лҸ„
- p.val: P-Value
лӢӨмқҢкіј к°ҷмқҙ мӮ¬мҡ©н•ңлӢӨ.
> source("normality.f")
> source("skew.f")
> source("kurtosis.f")
> cork.d <-read.table("cork.d", header=T)
> normality.f(cork.d)
Skewness is 4.476382 and P-value is 0.4036454 .
Don't reject the normality.
[edit]
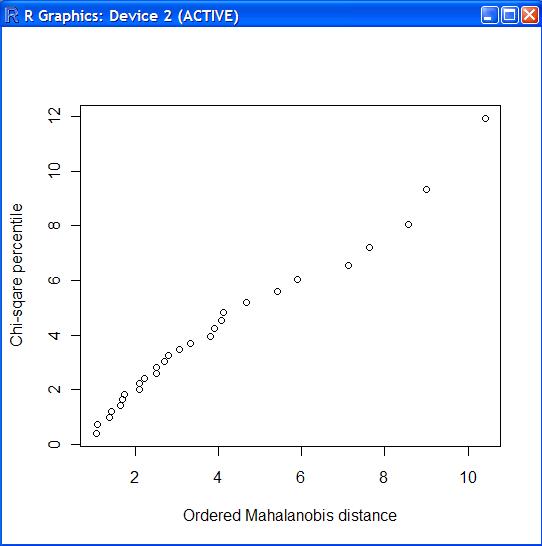
4 м№ҙмқҙм ңкіұ к·ёлҰј #
chiplot.f<-function(x)
{
n<-nrow(x)
p<-ncol(x)
mah<-sort(mahalanobis(x,mean(x),var(x)))
arr<-seq(from=1/(2*n),length=n,by=1/n);
chi.per<-qchisq(arr,p)
plot(mah,chi.per,xlab="Ordered Mahalanobis distance",ylab="Chi-sqare percentile");
}
> source("chiplot.f")
> chiplot.f(cork.d)
[edit]
5 лІ нғҖк·ёлҰј #
betaplot.f<-function(x)
{
n<-nrow(x)
p<-ncol(x)
alpha<-(p-2)/(2*p)
beta<-(n-p-3)/(2*(n-p-1))
mah<-sort((n/(n-1)^2)*mahalanobis(x,mean(x),var(x)))
start<-(1-alpha)/(n-alpha-beta+1)
step<-1/(n-alpha-beta+1)
arr<-seq(from=start,length=n,by=step)
beta.per<-qbeta(arr,alpha,beta)
plot(mah,beta.per,xlab="Ordered b_r",ylab="Beta percentile")
}
> source("betaplot.f")
> betaplot.f(cork.d)
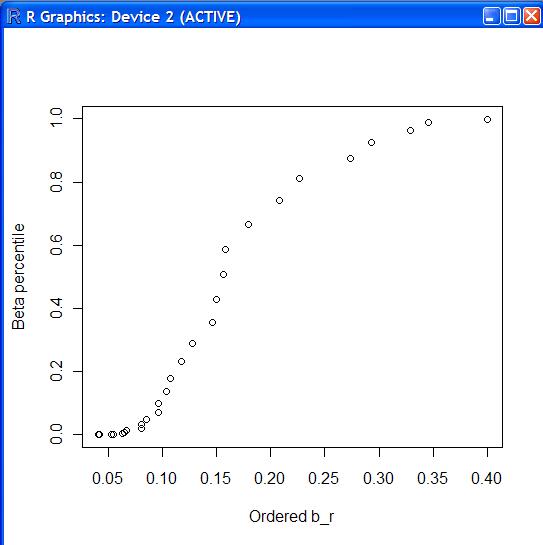
м •к·ңм„ұмқ„ к°Җм§Җкі мһҲмңјлӮҳ, кј¬лҰ¬ л¶Җ분мқҙ м•Ҫк°„ 짧мқҖ кІғмңјлЎң н•ҙм„қн• мҲҳ мһҲлӢӨ.
[edit]
6 Shapiro кІҖм • #
- к·Җл¬ҙк°Җм„Ө: м •к·ң분нҸ¬лҘј л”°лҘёлӢӨ. (p-value > 0.05)
- лҢҖлҰҪк°Җм„Ө: м •к·ң분нҸ¬лҘј л”°лҘҙм§Җ м•ҠлҠ”лӢӨ. (p-value < 0.05)
> source("rftn.f")
> rad.d <-read.table("rad.d", header=T)
> shapiro.test(rad.d$open)
Shapiro-Wilk normality test
data: rad.d$open
W = 0.8292, p-value = 1.977e-05
p-value < 0.05 мқҙлҜҖлЎң м •к·ң분нҸ¬лҘј л”°лҘҙм§Җ м•ҠлҠ”лӢӨ.
[edit]
7 Box-Cox Transformation #
м •к·ң 분нҸ¬м—җ к°Җк№қлҸ„лЎқ ліҖнҷҳн•ҳлҠ” л°©лІ•
mult.f<-function(lambda, x)
{
n<-nrow(x)
p<-ncol(x)
H<-rep(1,n)%*%t(rep(1,n))/n
Y<-matrix(0,n,p)
for(r in 1:n) {
for(i in 1:p) {
if( lambda[i] == 0 )
Y[r,i]<-log(x[r,i])
else
Y[r,i]<-(x[r,i]^lambda[i] - 1)/lambda[i]
}
}
Z<-matrix(0,n,p)
for(i in 1:p) {
a<-(prod(x[,i]))^((lambda[i]-1)/n)
Z[,i]<-Y[,i]/a
}
log(prod(eigen(t(Z)%*%(diag(n)-H)%*%Z)$values))
}
univ.f<-function(lambda, x)
{
n<-length(x)
H<-rep(1,n) %*% t(rep(1,n))/n
Y<-rep(0,n)
for(r in 1:n) {
if( lambda == 0 )
Y[r]<-log(x[r])
else
Y[r]<-(x[r]^lambda-1)/lambda
}
Y<-Y/(prod(x)^((lambda-1)/n))
t(Y)%*%(diag(n)-H)%*%Y
}
boxcox.f<-function(x)
{
x<-as.matrix(x)
p<-ncol(x)
sp<-rep(1,p)
for(i in 1:p) {
sp[i]<-nlminb(objective=univ.f,start=sp[i],x=x[,i])$par
}
lambda=nlminb(objective=mult.f,start=sp,x=x)$par
cat("Box-Cox parameters : ",lambda,"\n");
}
> source("boxcox.f")
> source("univ.f")
> source("mult.f")
> rad.d <-read.table("rad.d", header=T)
> chiplot.f(rad.d)
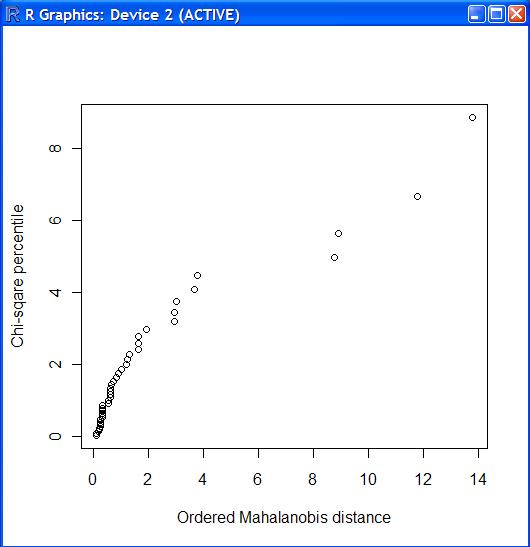
> boxcox.f(rad.d) Box-Cox parameters : 0.1607892 0.1511725
- lamda1 = 0.16
- lamda2 = 0.15
- closed = (closed ^ lamda1 - 1) / lamda1
- open = (open ^ lamda2 - 1) / lamda2
> boxcox_trans.d <- rad.d
> boxcox_trans.d$closed <- (rad.d$closed ^ 0.16 - 1) / 0.16
> boxcox_trans.d$open <- (rad.d$open ^ 0.15 - 1) / 0.15
> boxcox_trans.d
closed open
1 -1.6362440 -1.1015160
2 -1.9983371 -2.0210313
3 -1.4996718 -1.1015160
4 -1.9260564 -1.9470281
5 -2.3799621 -1.9470281
6 -1.7980629 -1.8161732
7 -2.0777107 -2.0210313
8 -2.3799621 -1.9470281
9 -2.0777107 -2.0210313
10 -1.9260564 -1.9470281
11 -2.1659062 -2.1928988
...
...
> chiplot.f(boxcox_trans.d)
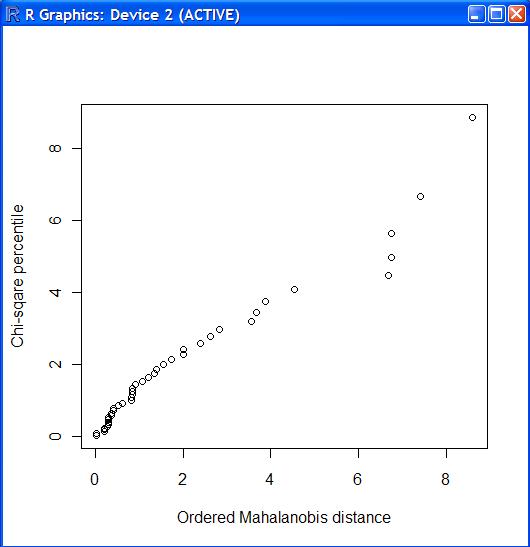
м•„лӢҲл©ҙ..
install.packages("TeachingDemos")
library("TeachingDemos")
set.seed(12345)
x <- rnorm(150,1,1)
e <- rnorm(150,0,2)
y <- (.6 *x + 13 + e)^3
hist(y)
hist(bct(y, 0.39))
shapiro.test(bct(y, 0.39))
мқҙл ҮкІҢ.. bctмқҳ 2лІҲм§ё л§Өк°ңліҖмҲҳ lamdaлҘј м Ғм ҲнһҲ мЎ°м Ҳ(0.1~0.9)н•ҙ к°Җл©ҙм„ң м •к·ң분нҸ¬лЎң л§Ңл“ лӢӨ..
п»ҝ